HEATSTER aims to develop a central platform for the work with heat stress transcription factors (HSFs). Currently, the platform provides a manually curated dataset that includes 848 plant HSF sequences
from 33 plant species (database version 1.0) and an extended dataset of 65 species (database version 2.0).
The extended dataset was identifed by the accessible annotation tool of this webserver. It allows the automatic identification and classification of HSFs.
Furthermore, a tool for visualization of HSF class motifs is provided. The dataset 1.0 of the HEATSTER database was curated by Lutz Nover and Klaus-Dieter Scharf, while the dataset version 2.0 was collected by Jannik Berz and Stefan Simm. The annotation tool was developed by Ingo Ebersberger and Sebastian Schuster.
The website and visualization tool was developed by Jannik Berz.
Critical comments and suggestions for further improvement are highly appreciated.
Refer to Stefan Simm or Ingo Ebersberger for bioinformatic aspects or HSF related questions.
Plant HSFs are encoded by multigene families consisting of 15 to 50 members. Based on conserved sequence and structural properties plant HSFs can be classified into three classes: HsfA, HsfB and HsfC. For each class, members can be further distinguished by forming subclasses indicated by numbers (e.g. HsfA1 vs. HsfA2), and multiple members within one subclass are differentiated by small case letters (e.g. HsfA1a vs. HsfA1b). An overview of the HSF families in the selected plant species and a comprehensive compilation of further details about plant HSF structure, function and evolution are reviewed by Scharf et al., 2012. The HSF names used in the database indicate the plant species abbreviated in a five letter code and the HSF-type. The Gene ID numbers refer to the gene/locus number used in the corresponding reference database.
The HEATSTER database contains the full length amino acid and open reading frame nucleotide sequences of heat stress transcription factors (HSFs) compiled for plant species with completed sequencing projects. The version 1.0 from January 2014 includes 32 manually curated angiosperm species with 26 eudicots and 6 monocots. The assignment of HSF sequences for database version 1.0 was performed on three levels:
Approximately 5 to 10% of all plant HSF sequence information available in databases like NCBI or Phytozome (until January 2014) turned out to be incomplete or incorrect annotated due to sequencing errors and/or wrongly predicted splicing sites. For this reason HEATSTER version 1.0 was created. HSF sequences in this database have been subjected to manual curation and artefacts introduced by sequencing or annotation errors had been removed to the best of our knowledge. The curation step was aided by homology comparison and scanning for conserved signature sequence motifs within the predicted genomic region and in the adjacent 5' and 3' flanking genome sequences. However, we are aware that in these cases our proposed annotation remains tentative until proven by experimental evidence.
HEATSTER version 1.0 should be used for the HSF classification of Monocotyledons and Eudicotyledons.
The version 1.0 harbours the essential functional motifs like DBD, OD, NLS, NES, AHA and RD as continuous signature sequence motifs.
HEATSTER version 2.0 from September 2016 extended the taxon compilation to 65 species including 1 Phaeophyta, 3 Rhodophyta and 61 species of the Viridiplantae, where the latter taxon comprises 5 Chlorophyta, 1 Bryophyta, 1 Lycopodiidae, 1 basal Magnoliophyta, 3 Gymnosperms, 16 Monocotyledons and 34 Eudicotyledons. In contrast to version 1.0, the HEATSTER HSF dataset version 2.0 was identified using the following procedure:
All sequences of the HEATSTER version 2.0 are directly extracted from the databases
Phytozome, NCBI, Dendrome
, Bambogdb, Banana hub,
Kazusa Database and Cucurbit Genomics Database (September 2016). Conserved signature motifs within the
predicted HSF sequences was performed by MEME and MAST (Timothy L. et al., 2009).
The version 2.0 is applicable for the HSF classification of all Viridiplantae species. Due to the usage of Chlorophyta, Gymnosperms and other plant ranks the motifs are more general due to phylogenetically more diverse training data. By this, approach the functional motifs like DBD, OD, NLS, NES, AHA and RD can be present as partial signature motifs in version 2.0. By this new functional motifs so far not mentioned can be assigned to HSF sequences.
While using the HEATSTER database for your research purposes, we recommend considering the rules associated with the use of the corresponding reference databases, especially in those cases where the genome sequencing projects are not published yet and the data are under protection of the Fort Lauderdale guidelines for large scale sequencing projects. We acknowledge the free accession to the genomic reference databases to build up the HSF database.
Conserved signature motifs within the predicted HSF sequences were performed by MEME, TOMTOM and MAST (Bailey et al. 2009). For each HSF sub-class (co-)orthologs of the selected plant species were used.
As classification of the sub-classes the widely accepted nomenclature of Nover et al. was used (Nover et al., 2001; Nover et al., 1996).
The HSF sub-classes were used to perform motif search via MEME (Bailey et al., 2009).
While for dataset version 1.0 a plain MEME search was conducted, dataset version 2.0 was created using different parameter sets for motif-width (10-20aa; 15-25aa; 20-30aa; 25-35aa;30-40aa) and site coverage (-OOPS; -ZOOPS; -Any).
Further, for each HSF sub-class we created a decoy database containing the random shuffled sequences of the HSF sub-class and perform the motif search ten times for each parameter setting.
The identification of the same signature motif in the decoy database was counted as false positive (FP) and used to calculate a false discovery rate (FDR).
All signature motifs below a threshold of 0.1 were selected as signature motifs for HSF sub-classes.
Further, signature motifs with an identity above 95% were merged via TOMTOM at the overlapping parts and extended to both endings.
Finally, the signature motifs of the single HSF sub-classes were cross-validated by searching the HSF sub-class signature motifs in the other HSF sub-class signature motif libraries with MAST to detect sub-class specific signature motifs.
While the signature library of dataset version 1.0 was created using only monocotyledons and eudicotyledons, the dataset of version 2.0 was created using all 65 species. For this reason,
version 1.0 might contain a more conserverd DBD and OD motif, while version 2.0 provides motifs conserved among all species and some smaller signature motifs for a
better discrimination between the HSF classes.
The HEATSTER annotation tool provides a platform that allows the automatic classification and annotation of unknown HSF sequences. The tool was developed on base of the motif
search tool MAST (Bailey et al., 1998) and includes optionally the possibility to analyze either a single amino acid sequence or a set of multiple sequences at the
same time. The classification and annotation is performed in two successive steps of repeated searches in a motif database, which was generated by using a training
set of selected HSF sequences for identification and classification of HSF class- and subclass-specific conserved amino acid sequence motifs, respectively.
The first step
of search determines the HSF class (A, B or C) to which the query sequence belongs. During the second step the particular HSF type within the identified class is assigned (sub-
classification, e.g. A1, A2,..., B1, B2...). As dataset of version 1.0 contains the more conserved DBD and OD motifs, this dataset is used for the classification.
The output of the single sequence analysis indicates the presence of the two most conserved HSF domains (DBD, OD with HR-A and HR-B region). The identification as HSF and classification of the HSF-type based on the similarity of the identified motifs compared to the corresponding consensus motifs is done based on the best e-value. For more detailed examination on the sequence level the alignment to the identified functional domains and subclass-specific sequence motifs can be visualized.
For analyzing more than one sequence at the same time the batch analysis pipeline can be used. This pipeline takes any number of sequences and allows the search for HSF sequences in more complex protein databases.
As MAST has only a limited capacity for searching in a sequence database with many sequences such as a whole proteome, the sequence database is filtered using a profile Hidden Markov Model (HMM) that describes the HSF-DBD domain.
Thereby, only sequences containing this domain are recognized as potential HSFs for further analysis, while all other sequences are discarded. The rest of the classification process is identical to the one with a single query sequence.
As output a table is provided, which shows all sequences classified as putative HSF with the indication of the presumable HSF type (class and subclass specification).
Further, a visualization is available for the HSF sequences, which indicates the motifs matched (motif set of version 1.0 and 2.0) onto the provided sequence.
The annotation tool relies on MAST (Timothy L. et al. , 1998) and HMMR (Finn et al., 2011).
The HEATSTER visualization tool allows comparative studies among HSFs and other sequences by analyzing their specific domain and motif composition. By this, it allows the user to detect signature motifs which might be specific for different HSF classes. Further, it is possible to detect differences between known classified HSFs and user defined sequences by matching their motif and domain composition.
During the analysis, a set of HSF class specific motifs is mapped onto the HSF sequences. Therefore, a plain and extended variant of the visualization tool is provided. In the plain version the user refers to HSF sequences from species, which are stored in the HEATSTER database version 2.0. In the extended version the user can submit a query sequence to compare this sequence to a set of different plant HSF classes. In both modes, the user is able to use the signature motif library of version 1.0, version 2.0 or both.
The visualisation tool relies
on a modified version of MAST (Bailey et al., 1998) from the MEME and MAST Suite.
Appearance and functionality of the output provided by the visualization tool is resembles the standard MAST output. Each sequence is represented as a continuous line. The matched signature motifs and domains are indicated as colorized boxes on top of the referring sequence. The colors are chosen randomly and independently for version 1.0 and 2.0. Thus, similar colors do not infer any relation between motifs within a version or between versions. By expanding the “Show/Hide” menu and by clicking on any colorized box, the sequence logo of the underlying motif shows up.
Further, for each sequence an e-value indicates the overall expectation to see chance all signature motifs of an Hsf class onto the sequence. The combined p-value indicates the overall p-value of all signature motifs matched onto the sequence. The visualization tool offers the option to display the single amino acids of the sequence by clicking the arrow beside the sequence name. By moving the gray boxes, the user can slice through the sequence. Start and stop position of the displayed sequence section are displayed inside the box. For each matched signature motif at a specific position the referring p-values is shown, and the sequence positions matching the motif are indicated by a + sign.
The HEATSTER contains HSF sequence information and classification of 33 plant species (version 1.0) and 65 species (version 2.0), respectively. As downloadable content it is possible to get all database information of HEATSTER version 1.0 or version 2.0. Deposited HSF-like and HSF-related lists are available as well as motif and domain sets of the different HSF sub-classes. The motifs for classification of the HSF are not necessarily the whole functional domain. Further a list of all analyzed plant species is deposited and contains a link to the downloaded proteome in the reference database. The proteomes were downloaded in April 2016.
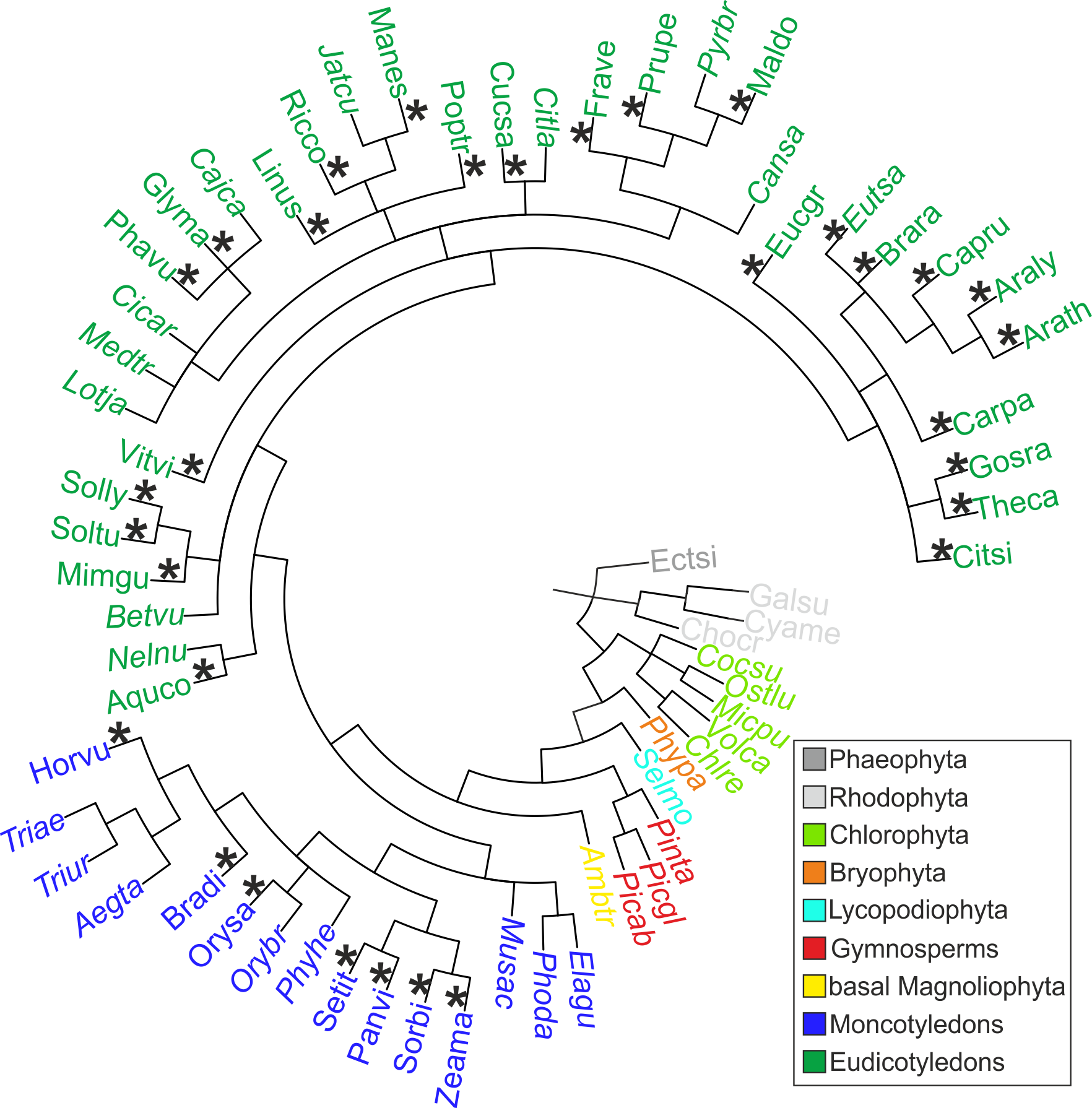
Figure: Species included in the HEATSTER databases. The taxonomic tree includes all proteomes of species included for the search of HSFs. In grey the Phaeophyta (dark grey) and Rhodophyta (light grey) are used as outgroups due to the fact, that they are not assigned to the VIridiplantae. All other species are color coded concerning their taxonomic rank. The HEATSTER database v 2.0 includes green algae (chlorophyta, light green), mosses (bryophyta, orange), club-mosses (lycopodiophyta, cyan), gymnosperms (red), basal mangoliophyta (yellow), monocots (blue) and eudicots (dark green). The species included in the visual curated HEATSTER database v 1.0 are marked with an asterisk and is limited to monocots and eudicots. The species are named concerning the five letter abbreviation (first three letters = genus and last two letters = species epithet) and the full list with their complete names can be downloaded in the HEATSTER data section.
Overview of plant species and reference databases:
| Name | Short name | Family | Data origin |
|---|---|---|---|
| Aegilops tauschii | Aegta | Monocotyledons | NCBI |
| Amborella trichopoda | Ambtr | basal Magnoliophyta | Phytozome |
| Aquilegia coerulea | Aquco | Eudicotyledons | Phytozome |
| Arabidopsis lyrata | Araly | Eudicotyledons | Phytozome |
| Arabidopsis thaliana | Arath | Eudicotyledons | Phytozome |
| Beta vulgaris | Betvu | Eudicotyledons | NCBI |
| Brachypodium distachyon | Bradi | Monocotyledons | Phytozome |
| Brassica rapa | Brara | Eudicotyledons | Phytozome |
| Cajanus cajan | Cajca | Eudicotyledons | NCBI |
| Cannabis sativa | Cansa | Eudicotyledons | NCBI |
| Capsella rubella | Capru | Eudicotyledons | Phytozome |
| Carica papaya | Carpa | Eudicotyledons | Phytozome |
| Chlamydomonas reinhardtii | Chlre | Chlorophyta | Phytozome |
| Chondrus crispus | Chocr | Rhodophyta | NCBI |
| Cicer arietinum | Cicar | Eudicotyledons | NCBI |
| Citrullus lanatus | Citla | Eudicotyledons | Cucurbit Genomics Database |
| Citrus sinensis | Citsi | Eudicotyledons | Phytozome |
| Coccomyxa subellipsoidea | Cocsu | Chlorophyta | Phytozome |
| Cucumis sativus | Cucsa | Eudicotyledons | Phytozome |
| Cyanidioschyzon merolae | Cyame | Rhodophyta | NCBI |
| Ectocarpus siliculosus | Ectsi | Phaeophyta | NCBI |
| Elaeis guineensis | Elagu | Monocotyledons | NCBI |
| Eucalyptus grandis | Eucgr | Eudicotyledons | Phytozome |
| Eutrema salsugineum | Eutsa | Eudicotyledons | Phytozome |
| Fragaria vesca | Frave | Eudicotyledons | Phytozome |
| Galdieria sulphuraria | Galsu | Rhodophyta | NCBI |
| Glycine max | Glyma | Eudicotyledons | Phytozome |
| Gossypium raimondii | Gosra | Eudicotyledons | Phytozome |
| Hordeum vulgare | Horvu | Monocotyledons | NCBI |
| Jatropha curcas | Jatcu | Eudicotyledons | NCBI |
| Linum usitatissimum | Linus | Eudicotyledons | Phytozome |
| Lotus japonicus | Lotja | Eudicotyledons | Kazusa Database |
| Malus domestica | Maldo | Eudicotyledons | Phytozome |
| Manihot esculenta | Manes | Eudicotyledons | Phytozome |
| Medicago truncatula | Medtr | Eudicotyledons | Phytozome |
| Micromonas pusilla | Micpu | Chlorophyta | Phytozome |
| Mimulus guttatus | Mimgu | Eudicotyledons | Phytozome |
| Musa acuminata | Musac | Monocotyledons | Banana hub |
| Musa balbisiana | Musba | Monocotyledons | Banana hub |
| Nelumbo nucifera | Nelnu | Eudicotyledons | NCBI |
| Oryza brachyantha | Orybra | Monocotyledons | NCBI |
| Oryza sativa | Orysa | Monocotyledons | Phytozome |
| Ostreococcus lucimarinus | Ostlu | Chlorophyta | Phytozome |
| Panicum virgatum | Panvi | Monocotyledons | Phytozome |
| Phaseolus vulgaris | Phavu | Eudicotyledons | Phytozome |
| Phoenix dactylifera | Phoda | Monocotyledons | NCBI |
| Phyllostachys heterocycla | Phyhe | Monocotyledons | Bamboogdb |
| Physcomitrella patens | Phypa | Bryophyta | Phytozome |
| Picea abies | Picab | Gymnosperms | Dendrome |
| Picea glauca | Picgl | Gymnosperms | Dendrome |
| Pinus taeda | Pnta | Gymnosperms | Dendrome |
| Populus trichocarpa | Poptr | Eudicotyledons | Phytozome |
| Prunus persica | Prupe | Eudicotyledons | Phytozome |
| Pyrus x bretschneideri | Pyrbr | Eudicotyledons | NCBI |
| Ricinus communis | Ricco | Eudicotyledons | Phytozome |
| Selaginella moellendorffii | Selmo | Lycopodiidae | Phytozome |
| Setaria italica | Setit | Monocotyledons | Phytozome |
| Solanum lycopersicum | Solly | Eudicotyledons | Phytozome |
| Solanum tuberosum | Soltu | Eudicotyledons | Phytozome |
| Sorghum bicolor | Sorbi | Monocotyledons | Phytozome |
| Thellungiella halophila | Thelha | Eudicotyledons | Phytozome |
| Theobroma cacao | Theca | Eudicotyledons | Phytozome |
| Triticum aestivum | Triae | Monocotyledons | Phytozome |
| Triticum urartu | Triur | Monocotyledons | NCBI |
| Vitis vinifera | Vitvi | Eudicotyledons | Phytozome |
| Volvox carteri | Volca | Chlorophyta | Phytozome |
| Zea mays | Zeama | Monocotyledons | Phytozome |
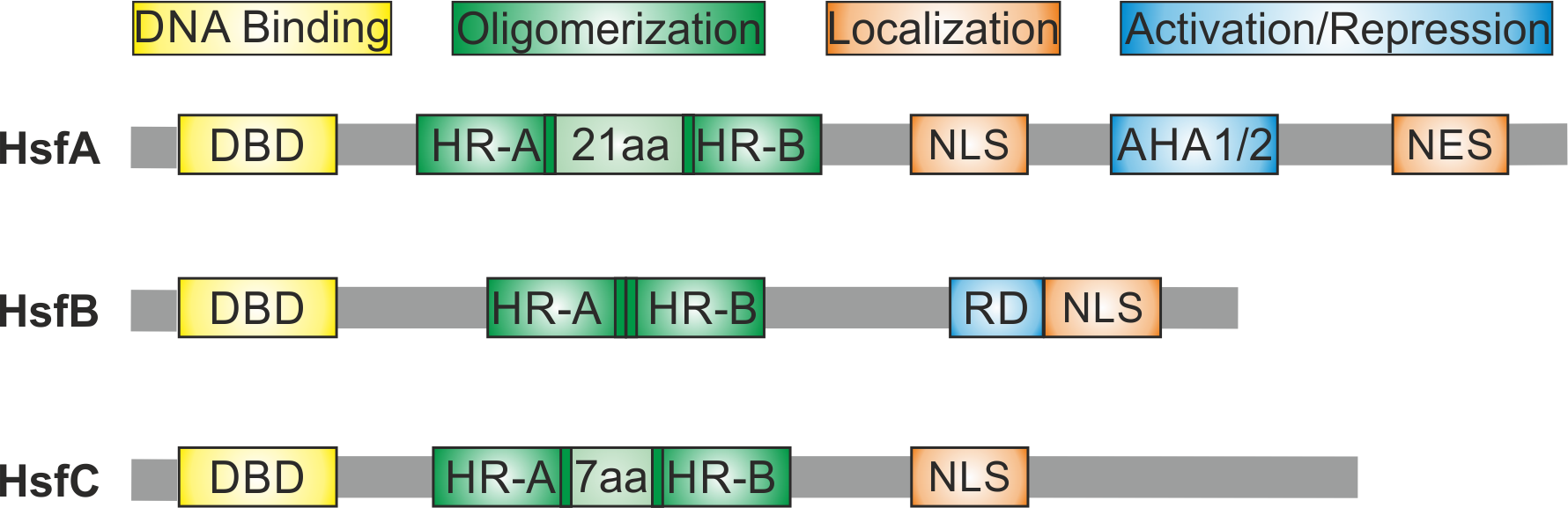
DBD: HSF DNA-binding domain with the central and mostly conserved winged helix-turn-helix motif
OD: HSF oligomerization domain, formation of homo- and hetero-oligomeric HSF complexes
HR-A/B: heptad pattern of hydrophobic amino acid (aa) residues forming the OD, the N-terminal region (part A) is separated from the C-terminal part B by a linker region of 6 aa residues in class B HSFs, and additionally inserted 21 aa residues in class A or 7 aa residues in class C HSFs
NLS: HSF nuclear localization sequence, monopartite (m) or bipartite (b)
NES: HSF nuclear export sequence
AHA: HSF transcription activator motifs, consisting of aromatic (A), large hydrophobic (H) and acidic (A) aa residues, typically identified in class A HSFs
RD: HSF transcription repression domain, identified in distinct class B HSFs
When using HEATSTER please cite:
Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim Biophys Acta (2012) 1819:104-119, doi:10.1016/j.bbagrm.2011.10.002
For HSF annotation, please use the widley accepted nomenclature suggested in:
Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim Biophys Acta (2012) 1819:104-119, doi:10.1016/j.bbagrm.2011.10.002
Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim Biophys Acta (2012) 1819:104-119, doi:10.1016/j.bbagrm.2011.10.002
Bailey, T.L., et al. (2009) MEME SUITE: tools for motif discovery and searching, Nucleic acids research, 37, W202-208.
Finn, R.D., Clements, J. and Eddy, S.R. (2011) HMMER web server: interactive sequence similarity searching, Nucleic acids research, 39, W29-37.
Nover, L., et al. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need?, Cell Stress Chaperon, 6, 177-189.
Nover, L., et al. (1996) The Hsf world: classification and properties of plant heat stress transcription factors, Cell Stress Chaperones, 1, 215-223.
Further, Bootstrap, jQuery, and sorttable were used. The underlying HSF database relies on MySQL. The website is designed using HTML, CSS and PHP and annotation tool is writte in Perl.
This site is hosted and maintained by:
Critical comments and suggestions for further improvement are highly appreciated. Refer to Stefan Simm or to Ingo Ebersberger for HSF-related queries and bioinformatics aspects.
For acknowledging the use of the HEATSTER platform please refer to:
Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim Biophys Acta (2012) 1819:104-119, doi:10.1016/j.bbagrm.2011.10.002
For HSF annotation, please use the widley accepted nomenclature suggested in:
Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim Biophys Acta (2012) 1819:104-119, doi:10.1016/j.bbagrm.2011.10.002
This file is part of HEATSTER database.
HEATSTER database is free software: you can redistribute it and/or modify
it under the terms of the GNU General Public License as published by
the Free Software Foundation, either version 3 of the License, or
(at your option) any later version.
HEATSTER database is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with HEATSTER database. If not, see GNU licence.
For usage and license of included tools, please refer to MEME Suite and HMMER. While using the HEATSTER database for your research purposes we recommend considering the rules associated with the use of the corresponding reference databases, especially in those cases where the genome sequencing projects are not published yet and the data are under protection of the Fort Lauderdale guidelines for large scale sequencing projects.
Data privacy statement
By using our website you consent to the collection, processing and use of data as described below. Our website can be visited without registration. This data such as pages called or name of the retrieved file, date and time are stored on the server for statistical purposes, without direct relation of this data to your person. Personal data, in particular name, address or e-mail address are collected as far as possible on a voluntary basis. Without your consent, the data will not be passed on to third parties.
Privacy policy for cookies
Currently, our website is not using cookies.
Source: Data privacy sample
Homepage: 2.0
Database: version 1.0 and 2.0 included.
Refer to:
Stefan Simm
or
Ingo Ebersberger